Preamble
In Momocs, a shape is a matrix of (x, y) coordinates. When they are plenty, they are usually gathered in a Coo object which is, essentially, a list with at least a $coo component.
## [1] TRUE## [,1] [,2]
## [1,] 37 561
## [2,] 40 540
## [3,] 40 529
## [4,] 43 508
## [5,] 46 487
## [6,] 48 477## [1] "Out" "Coo"## [1] TRUE## [1] "list"## $brahma
## [,1] [,2]
## [1,] 37 561
## [2,] 40 540
## [3,] 40 529
## [4,] 43 508
## [5,] 46 487
## [6,] 48 477
##
## $caney
## [,1] [,2]
## [1,] 53 535
## [2,] 53 525
## [3,] 54 505
## [4,] 53 495
## [5,] 54 485
## [6,] 54 464Most operations on shapes can also be done on Coo objects. For example, getting the area or centering:
## [1] 234515## brahma caney chimay corona deusventrue duvel
## 234515.0 201056.5 119459.5 119568.5 165735.5 114015.0In other words most coo_ functions are actually methods that can actually be passed with a single matrix or a Coo object.
Below, we will distinguish coo_* operations based on whether they return a shape (eg if it is a geometric operation) or a scalar (a single number that can be used as a shape descriptor).
Not all are described but you can obtain a full list of coo_ operations with:
## [1] "coo_align" "coo_aligncalliper" "coo_alignminradius"
## [4] "coo_alignxax" "coo_angle_edges" "coo_angle_tangent"## [1] 95Scalar descriptors of shape
coo_scalar calculate all scalar descriptors included in Momocs. coo_rectilinearity is not included by default since it takes a looot of time to compute.
## # A tibble: 30 x 15
## area calliper centsize circularity circularityharali… circularitynorm
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 20030. 319. 87.9 54.4 2.21 4.33
## 2 26078. 311. 103. 32.0 2.47 2.55
## 3 31516 280. 91.2 51.1 2.43 4.07
## 4 14180. 231. 73.0 46.6 2.84 3.71
## 5 9282. 206. 63.4 41.8 2.33 3.32
## 6 24060. 263. 91.2 35.9 3.84 2.85
## 7 14940. 263. 81.5 52.0 2.74 4.14
## 8 33921 352. 109. 35.7 2.83 2.84
## 9 18654 209. 67.3 74.1 2.45 5.89
## 10 36654. 312. 98.3 106. 2.72 8.45
## # ... with 20 more rows, and 9 more variables: convexity <dbl>,
## # eccentricityboundingbox <dbl>, eccentricityeigen <dbl>,
## # elongation <dbl>, length <dbl>, perim <dbl>, rectangularity <dbl>,
## # solidity <dbl>, width <dbl>Each of the function listed below has its own method that you can used by prefixing it with coo_; see the example below with coo_area.
## area
## calliper
## centsize
## circularity
## circularityharalick
## circularitynorm
## convexity
## eccentricityboundingbox
## eccentricityeigen
## elongation
## length
## perim
## rectangularity
## solidity
## width## [1] 14180.5## arrow bone buttefly cat check cross dog fish
## 20029.5 26078.5 31516.0 14180.5 9281.5 24060.5 14939.5 33921.0
## hand hands heart info lady leaf leaf2 leaf3
## 18654.0 36653.5 24276.5 8453.5 11541.0 12861.5 16993.5 18301.5
## leaf4 moon morph parrot penta pigeon plane puzzle
## 17748.5 13558.0 28292.0 7894.5 24709.0 23792.5 12649.0 16986.5
## rabbit sherrif snail star tetra umbrella
## 18915.5 15766.0 26992.5 23132.0 18119.0 16878.5## # A tibble: 6 x 2
## area perim
## <dbl> <dbl>
## 1 20030. 1044.
## 2 26078. 914.
## 3 31516 1269.
## 4 14180. 813.
## 5 9282. 623.
## 6 24060. 929.Geometric operations
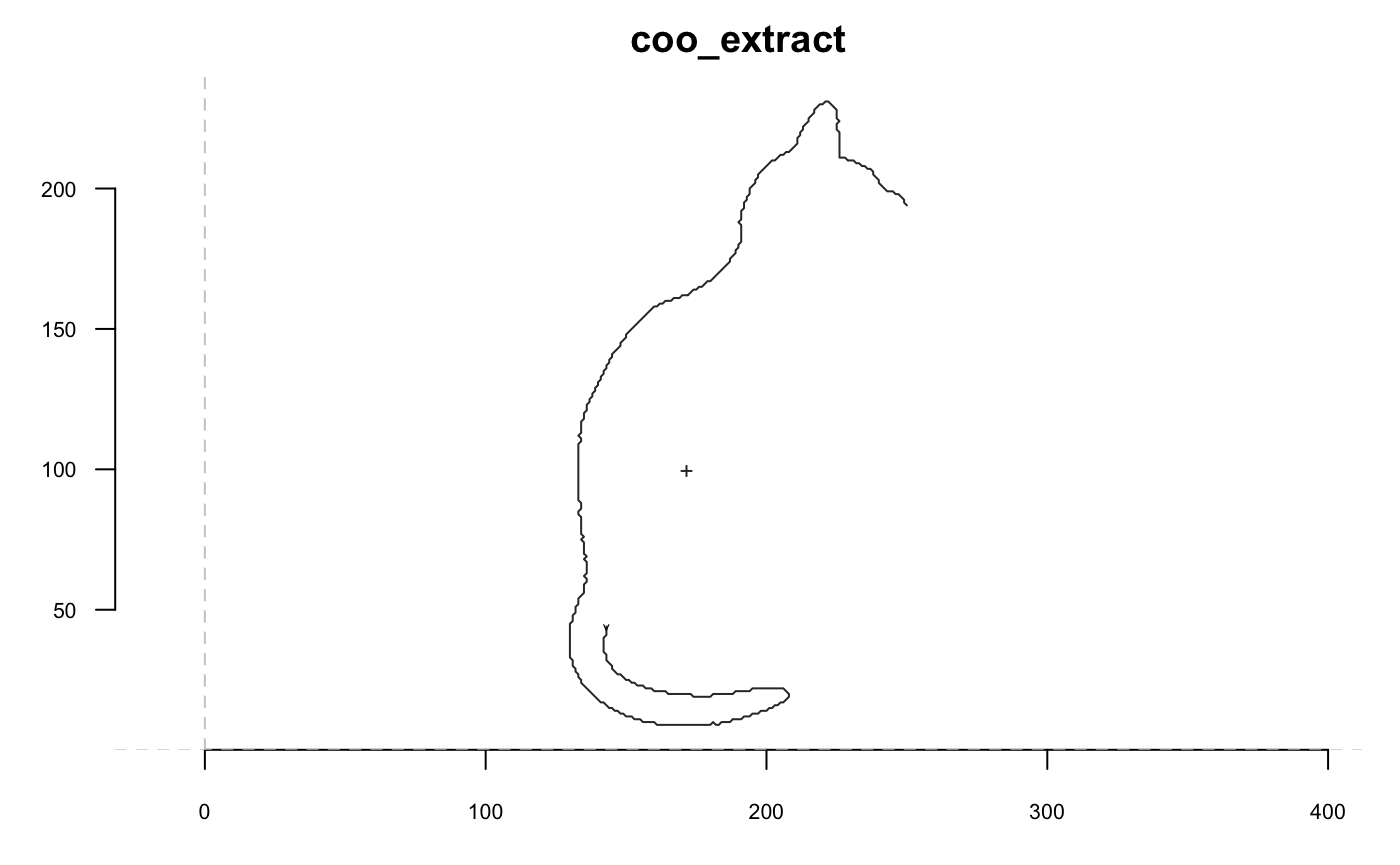
The functions below return shapes, so we exemplify them with graphs. For the sake of clarity/speed we illustrate them on single shape but, again, this can be done on Coo objects.
## will soon be deprecated, see ?pile
## will soon be deprecated, see ?pile
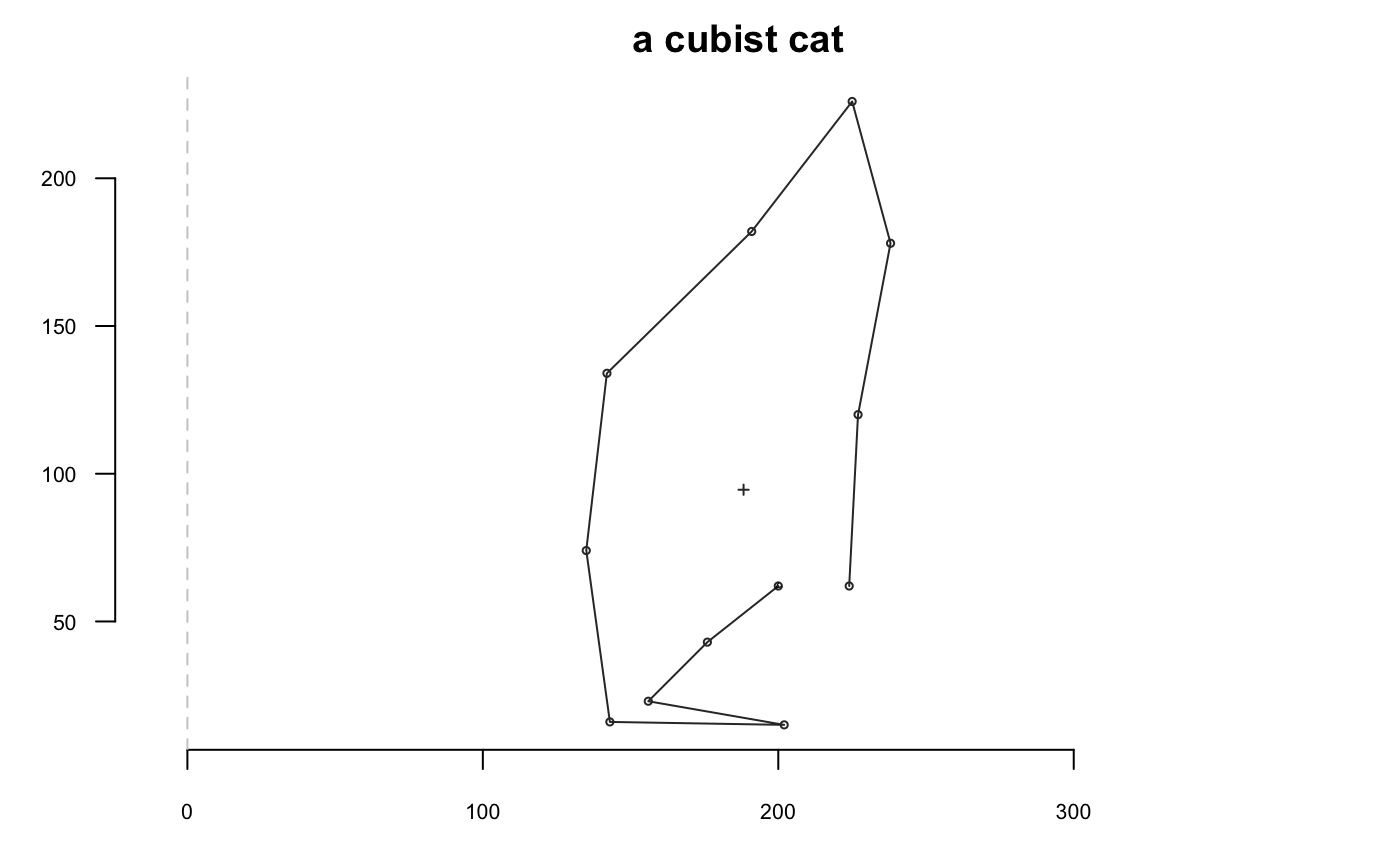
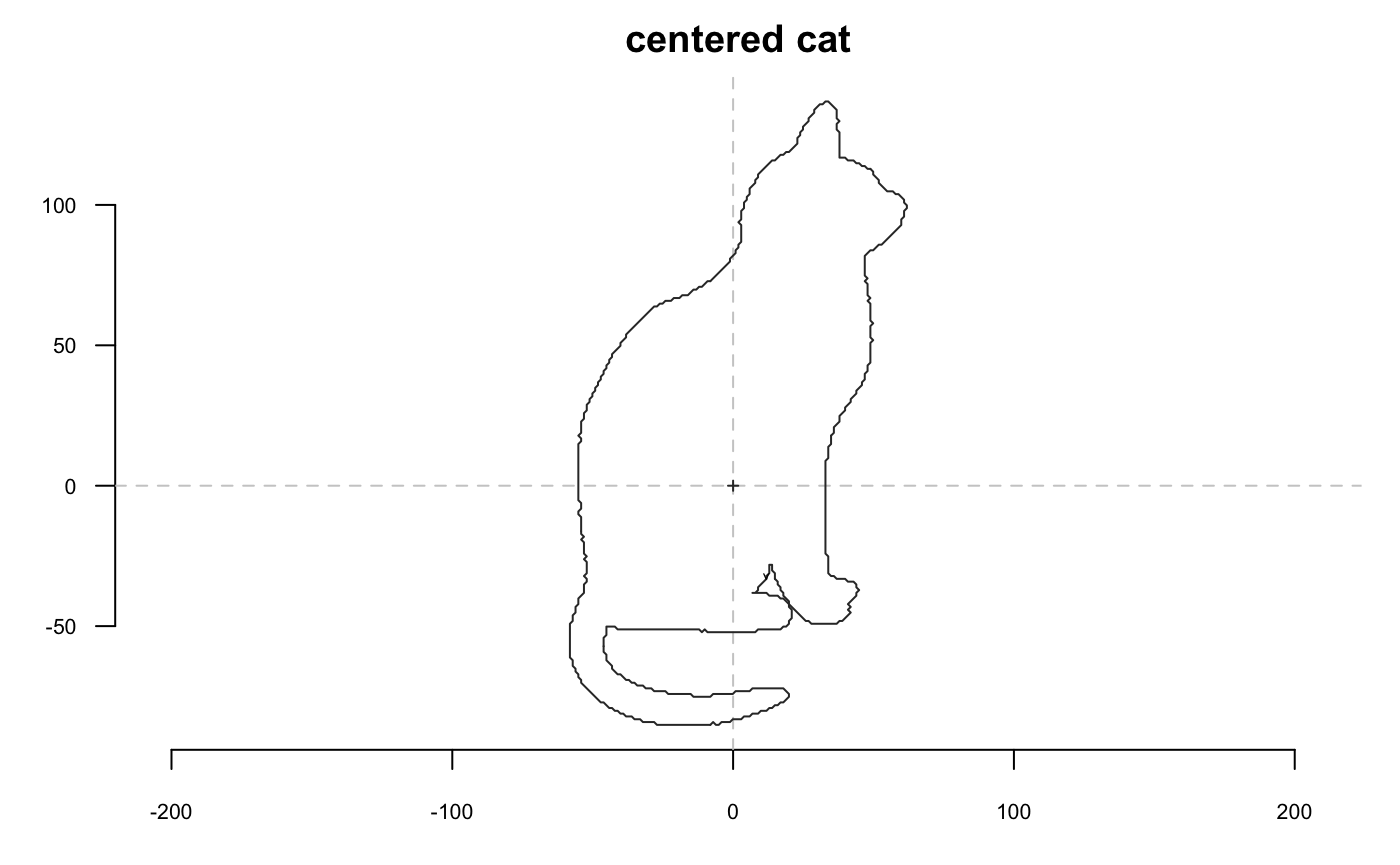
Now we go with a cat and we will extensively use coo_plot. We define a function that will help display side by side the original and transformed cat; when I use p(coo_align), it is equivalent to coo_align(your_shp) or shp %>% coo_align
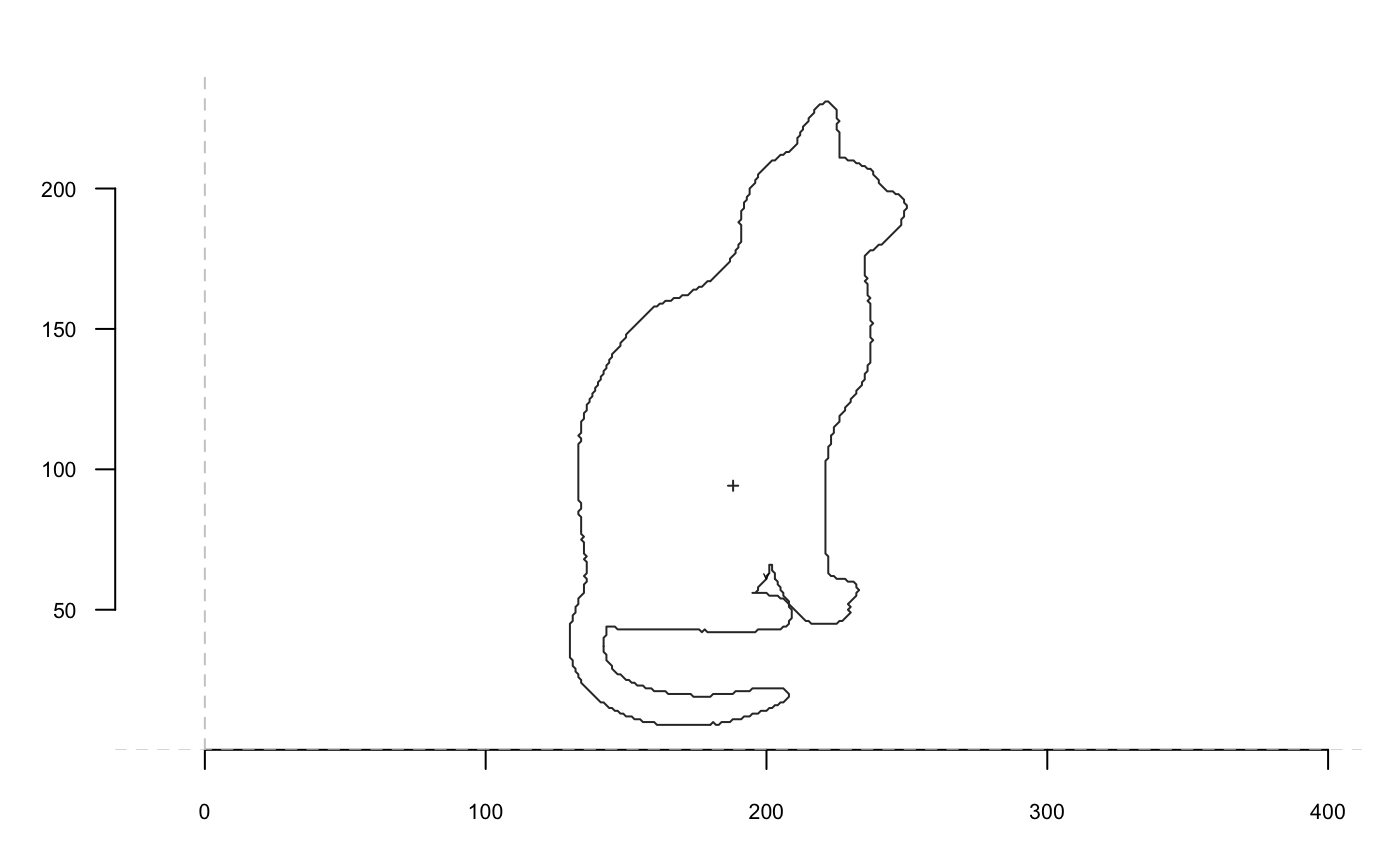
Size
Rotation
shp %>% coo_rotatecenter(-pi/6, center=c(250, 195)) %>% coo_plot(main="coo_rotatecenter") # with center ~on cat's nose
Sampling
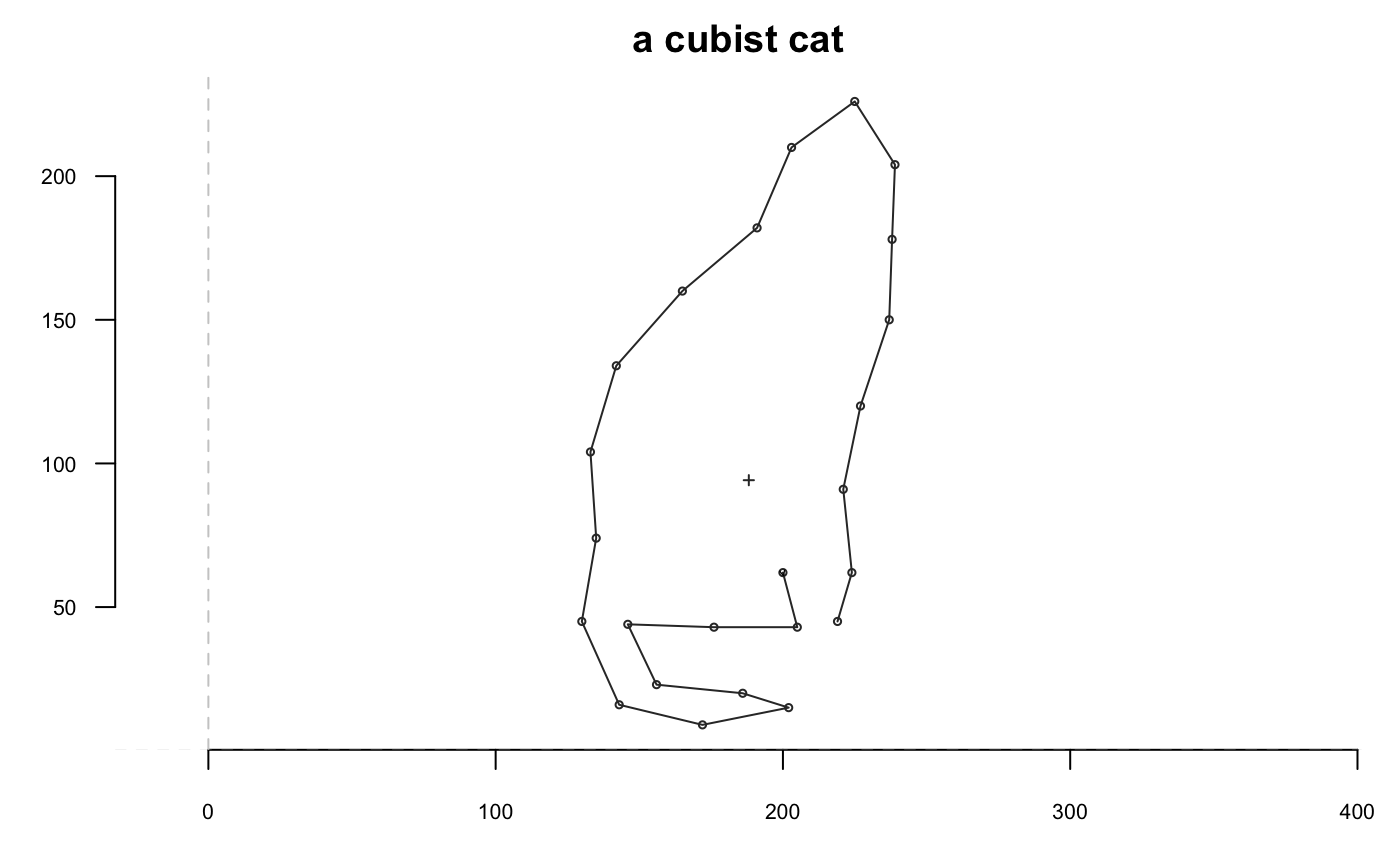
## [1] 710shp %>% coo_sample(36) %>% coo_plot(main="coo_sample")
shp %>% coo_sample_prop(1/20) %>% coo_plot(main="coo_sample")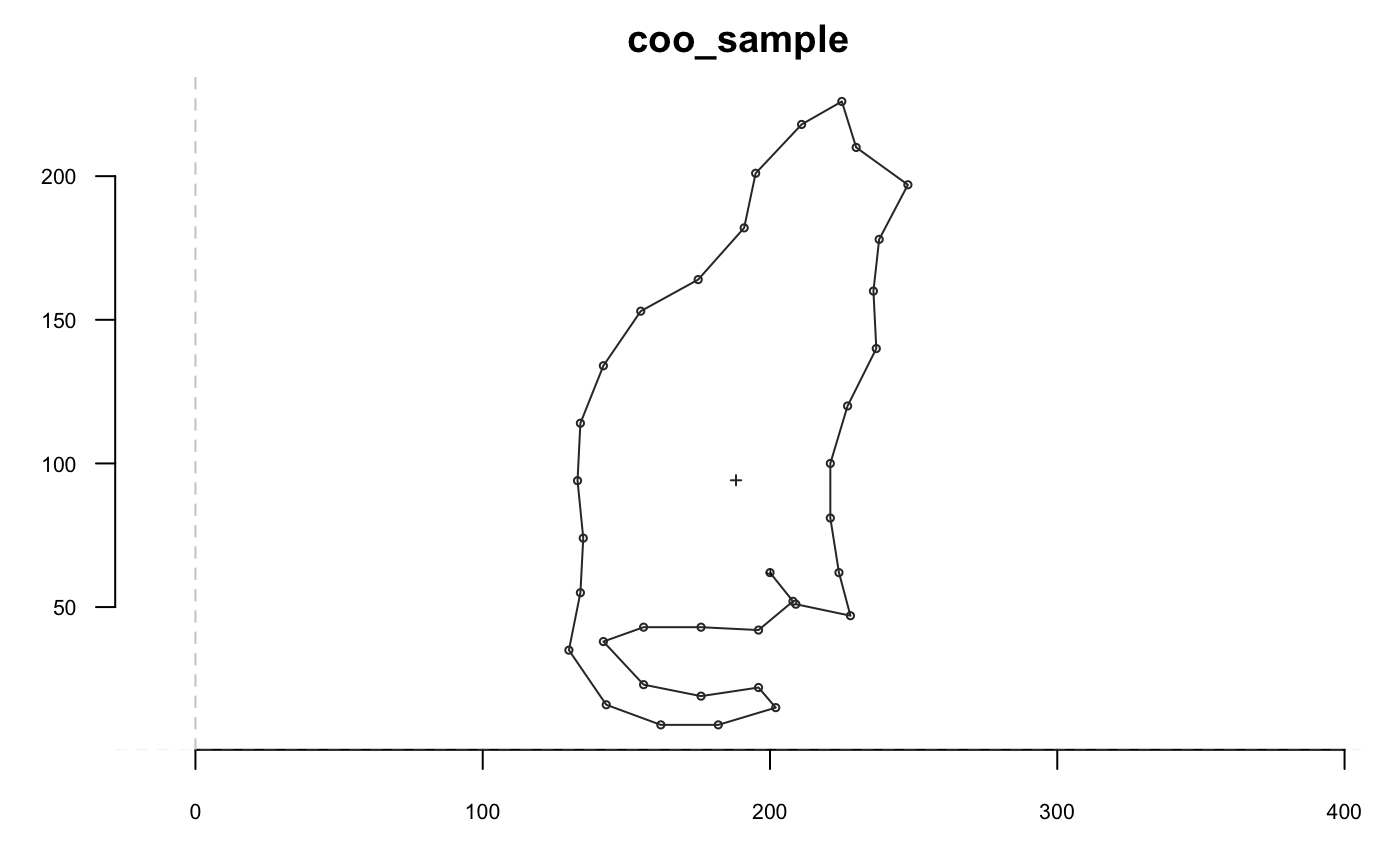
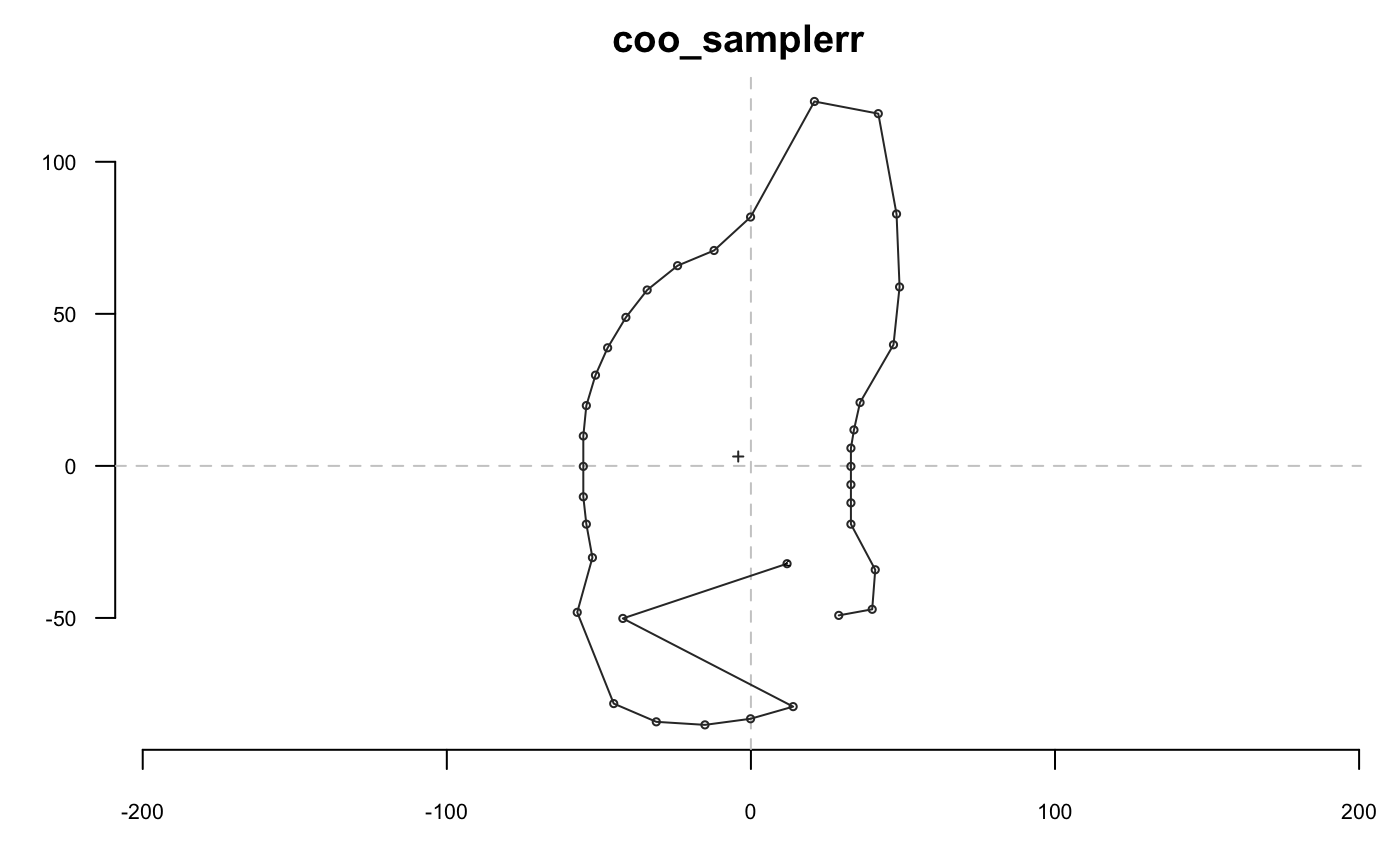
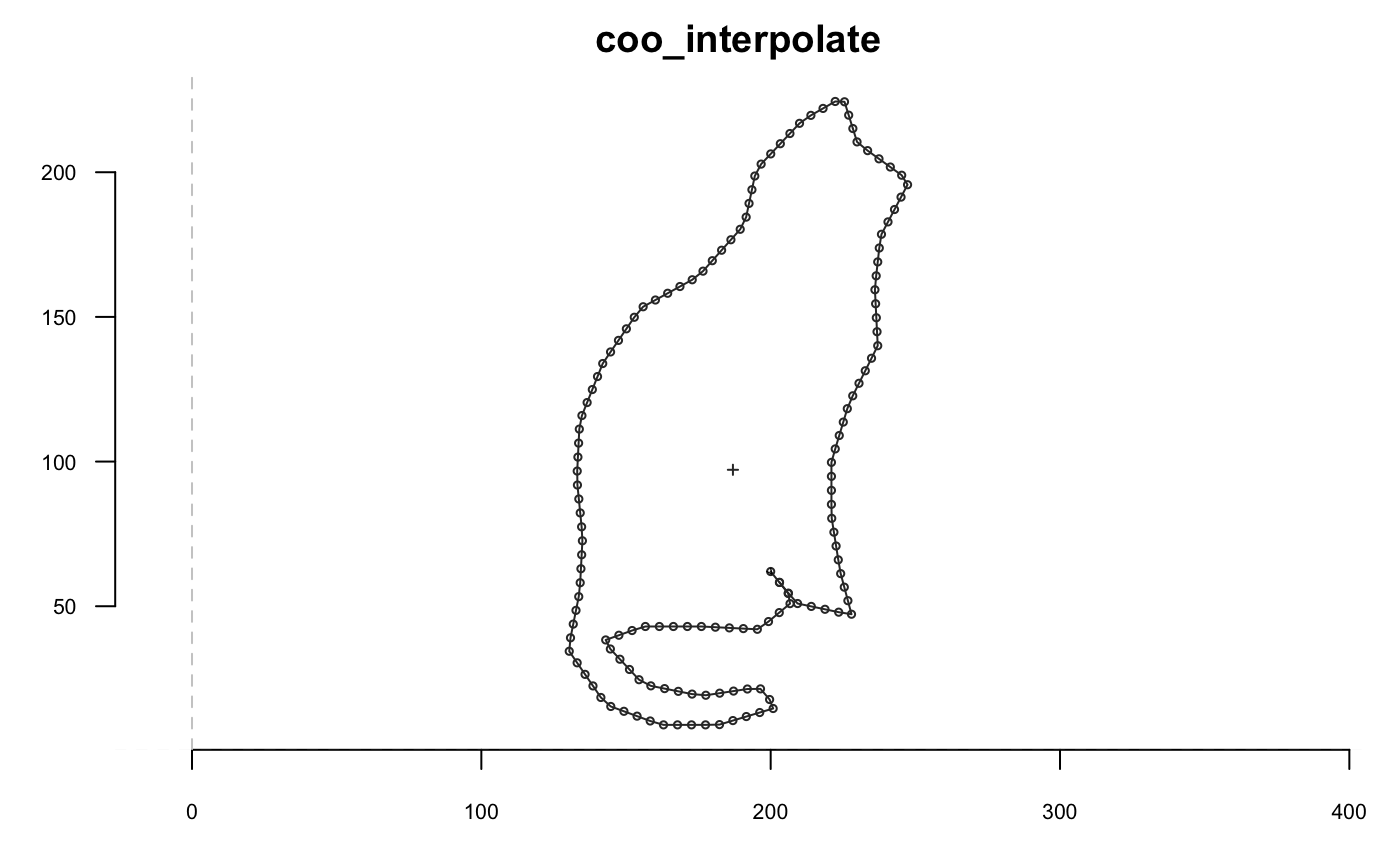
Slicing
Based on geometry
 If you want to slice but want the first point to be on one end, then
If you want to slice but want the first point to be on one end, then coo_slidegap is your friend :
Indexing
Sliding
shp %>% coo_slide(id=93) %>% coo_plot(main="coo_slide", cex.first.point = 2) # same comment for ldk/Coo as for coo_slice
Misc
A side effect of coo_dxy is to set the first point on (0, 0)
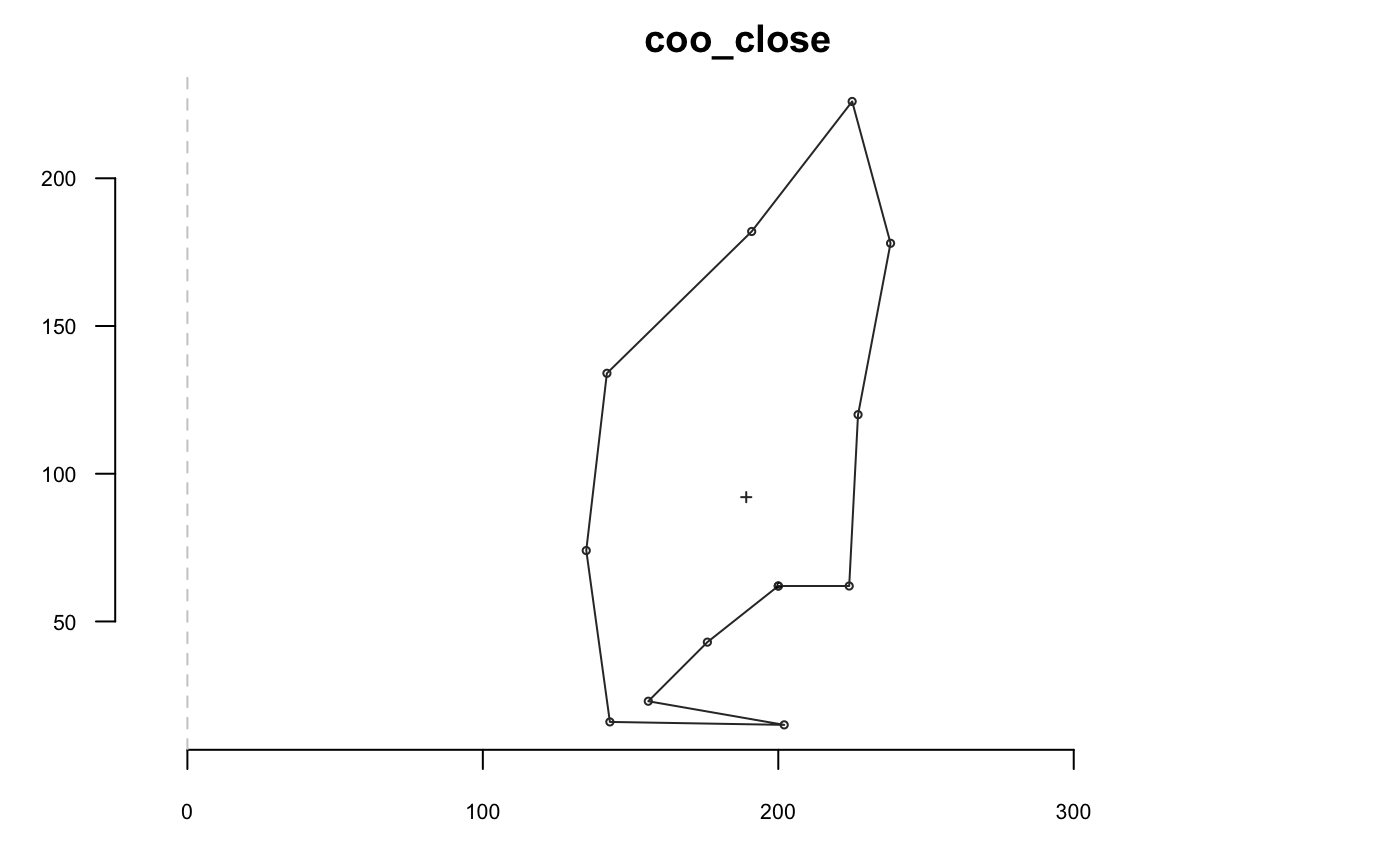
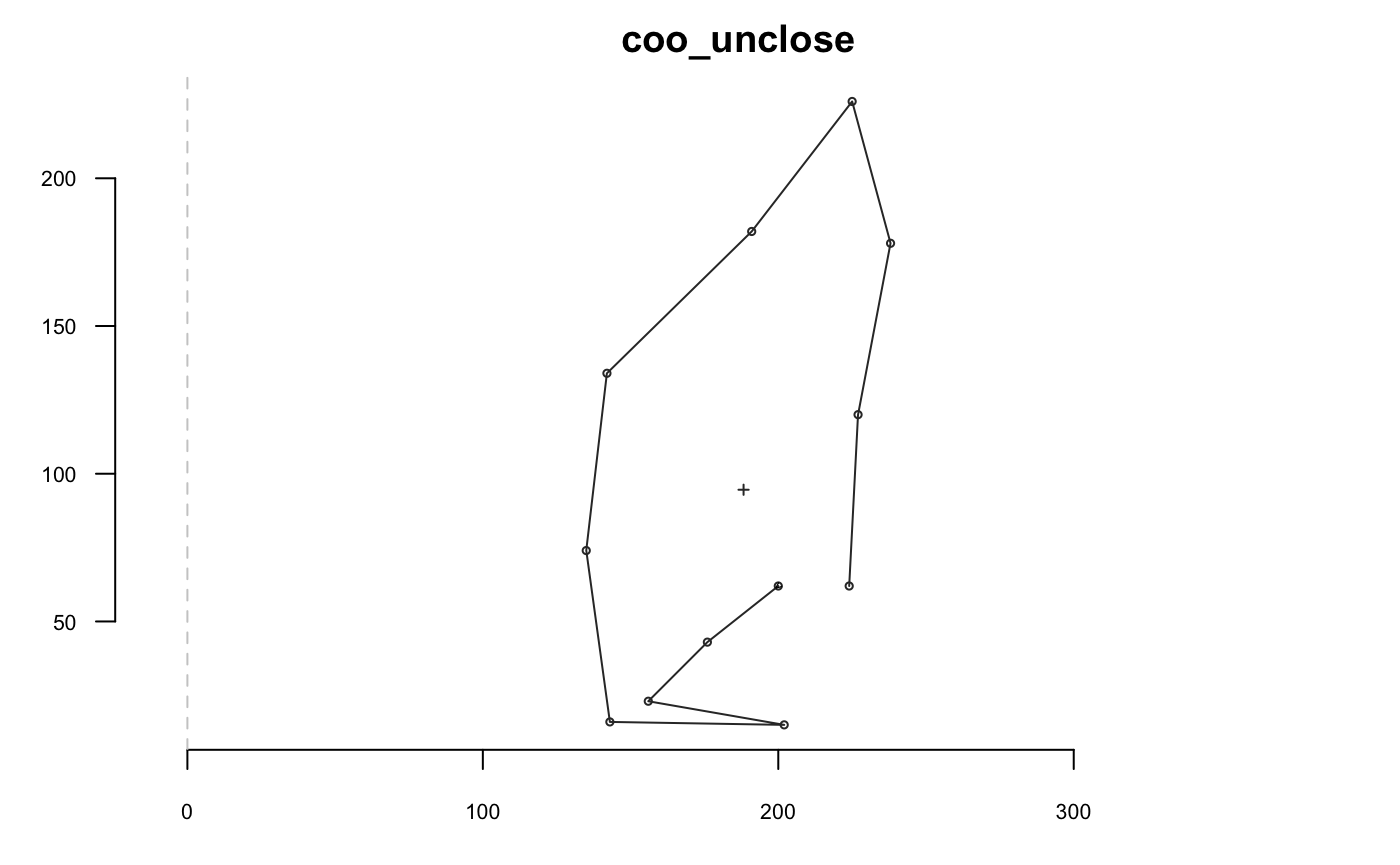
coo_force2close can be use to spread differences between the first and last point, and to force closing:
# for the sake of reproducibility
set.seed(123)
shp %>% coo_jitter(factor=10) %>% coo_plot(main="coo_jitter")
Other coo operations
# coo_descriptors along
coo_centdist
coo_perimcum
coo_perimpts
# coo_descriptors non-scalars
coo_boundingbox
coo_centpos # should be a df?
coo_chull
coo_chull_onion
coo_diffrange # coo_range_diff
coo_lw
coo_truss
coo_range
coo_range_enlarge
# coo_drawers
coo_arrows
coo_draw
coo_draw_rads
coo_listpanel
coo_lolli
coo_oscillo # deprecate for a proper oscillo
coo_plot
coo_ruban
# coo_others
coo_angle_edges
coo_angle_tangent
coo_intersect_angle
coo_intersect_direction
coo_intersect_segment
# coo_testers
coo_is_closed
coo_likely_anticlockwise
coo_likely_clockwise
# helpers
coo_check
coo_nb
coo_ldk