A very basic implementation of k-means. Beware that morphospaces are calculated so far for the 1st and 2nd component.
Usage
KMEANS(x, ...)
# S3 method for class 'PCA'
KMEANS(x, centers, nax = 1:2, pch = 20, cex = 0.5, ...)Arguments
- x
PCA object
- ...
additional arguments to be passed to kmeans
- centers
numeric number of centers
- nax
numeric the range of PC components to use (1:2 by default)
- pch
to draw the points
- cex
to draw the points
Value
the same thing as kmeans
See also
Other multivariate:
CLUST(),
KMEDOIDS(),
LDA(),
MANOVA(),
MANOVA_PW(),
MDS(),
MSHAPES(),
NMDS(),
PCA(),
classification_metrics()
Examples
data(bot)
bp <- PCA(efourier(bot, 10))
#> 'norm=TRUE' is used and this may be troublesome. See ?efourier #Details
KMEANS(bp, 2)
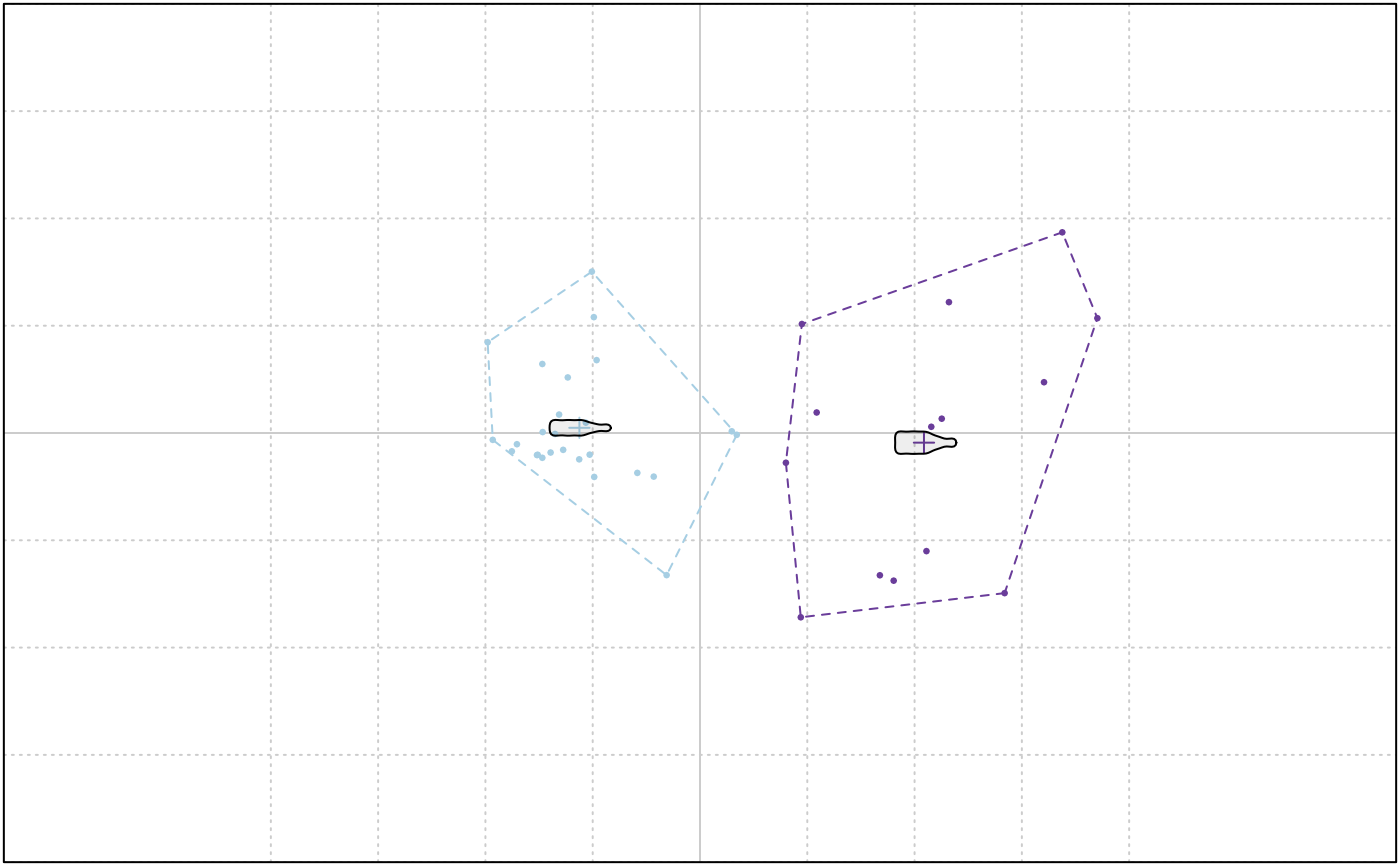 #> K-means clustering with 2 clusters of sizes 14, 26
#>
#> Cluster means:
#> PC1 PC2
#> 1 0.07496282 -0.003229186
#> 2 -0.04036460 0.001738792
#>
#> Clustering vector:
#> brahma caney chimay corona deusventrue
#> 2 2 1 2 2
#> duvel franziskaner grimbergen guiness hoegardeen
#> 1 2 1 2 2
#> jupiler kingfisher latrappe lindemanskriek nicechouffe
#> 2 2 1 2 2
#> pecheresse sierranevada tanglefoot tauro westmalle
#> 2 1 1 2 2
#> amrut ballantines bushmills chivas dalmore
#> 2 1 2 1 1
#> famousgrouse glendronach glenmorangie highlandpark jackdaniels
#> 2 2 2 1 2
#> jb johnniewalker magallan makersmark oban
#> 2 2 2 1 2
#> oldpotrero redbreast tamdhu wildturkey yoichi
#> 1 1 2 2 1
#>
#> Within cluster sum of squares by cluster:
#> [1] 0.03758606 0.02127484
#> (between_SS / total_SS = 67.3 %)
#>
#> Available components:
#>
#> [1] "cluster" "centers" "totss" "withinss" "tot.withinss"
#> [6] "betweenss" "size" "iter" "ifault"
#> K-means clustering with 2 clusters of sizes 14, 26
#>
#> Cluster means:
#> PC1 PC2
#> 1 0.07496282 -0.003229186
#> 2 -0.04036460 0.001738792
#>
#> Clustering vector:
#> brahma caney chimay corona deusventrue
#> 2 2 1 2 2
#> duvel franziskaner grimbergen guiness hoegardeen
#> 1 2 1 2 2
#> jupiler kingfisher latrappe lindemanskriek nicechouffe
#> 2 2 1 2 2
#> pecheresse sierranevada tanglefoot tauro westmalle
#> 2 1 1 2 2
#> amrut ballantines bushmills chivas dalmore
#> 2 1 2 1 1
#> famousgrouse glendronach glenmorangie highlandpark jackdaniels
#> 2 2 2 1 2
#> jb johnniewalker magallan makersmark oban
#> 2 2 2 1 2
#> oldpotrero redbreast tamdhu wildturkey yoichi
#> 1 1 2 2 1
#>
#> Within cluster sum of squares by cluster:
#> [1] 0.03758606 0.02127484
#> (between_SS / total_SS = 67.3 %)
#>
#> Available components:
#>
#> [1] "cluster" "centers" "totss" "withinss" "tot.withinss"
#> [6] "betweenss" "size" "iter" "ifault"
