Usage
LDA(x, fac, retain, ...)
# Default S3 method
LDA(x, fac, retain, ...)
# S3 method for class 'PCA'
LDA(x, fac, retain = 0.99, ...)Arguments
- x
a Coe or a PCA object
- fac
the grouping factor (names of one of the $fac column or column id)
- retain
the proportion of the total variance to retain (if retain<1) using scree, or the number of PC axis (if retain>1).
- ...
additional arguments to feed lda
Value
a 'LDA' object on which to apply plot.LDA, which is a list with components:
xany Coe object (or a matrix)facgrouping factor usedremovedids of columns in the original matrix that have been removed since constant (if any)modthe raw lda mod from ldamod.predthe predicted model using x and modCV.faccross-validated classificationCV.tabcross-validation tabkeCV.correctproportion of correctly classified individualsCV.ceclass errorLDsunstandardized LD scores see Claude (2008)mshapemean values of coefficients in the original matrixmethodinherited from the Coe object (if any)
Note
For LDA.PCA, retain can be passed as a vector (eg: 1:5, and retain=1, retain=2, ..., retain=5) will be tried, or as "best" (same as before but retain=1:number_of_pc_axes is used).
Silent message and progress bars (if any) with options("verbose"=FALSE).
See also
Other multivariate:
CLUST(),
KMEANS(),
KMEDOIDS(),
MANOVA(),
MANOVA_PW(),
MDS(),
MSHAPES(),
NMDS(),
PCA(),
classification_metrics()
Examples
bot.f <- efourier(bot, 24)
#> 'norm=TRUE' is used and this may be troublesome. See ?efourier #Details
bot.p <- PCA(bot.f)
LDA(bot.p, 'type', retain=0.99) # retains 0.99 of the total variance
#> 8 PC retained
#> * Cross-validation table ($CV.tab):
#> classified
#> actual beer whisky
#> beer 17 3
#> whisky 1 19
#>
#> * Class accuracy ($CV.ce):
#> beer whisky
#> 0.85 0.95
#>
#> * Leave-one-out cross-validation ($CV.correct): (90% - 36/40):
LDA(bot.p, 'type', retain=5) # retain 5 axis
#> 5 PC retained
#> * Cross-validation table ($CV.tab):
#> classified
#> actual beer whisky
#> beer 16 4
#> whisky 4 16
#>
#> * Class accuracy ($CV.ce):
#> beer whisky
#> 0.8 0.8
#>
#> * Leave-one-out cross-validation ($CV.correct): (80% - 32/40):
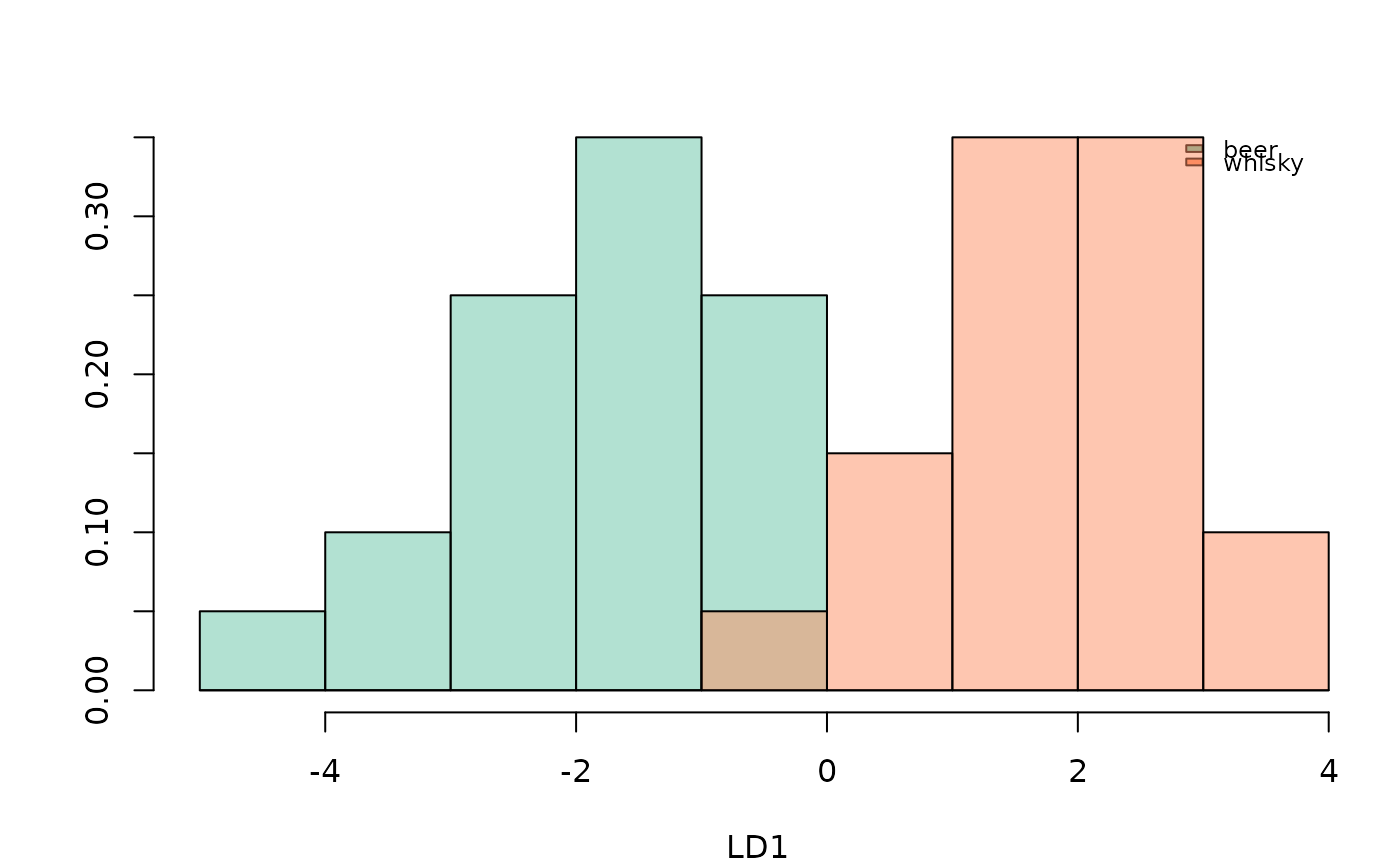
bot.l <- LDA(bot.p, 'type', retain=0.99)
#> 8 PC retained
plot_LDA(bot.l)
#> * Only two levels, so a single LD and preparing for an histogram
 #> $xy
#> LD1
#> brahma 2.06882655
#> caney 1.95733171
#> chimay 3.18567319
#> corona 1.91972111
#> deusventrue 1.51983847
#> duvel 3.25459981
#> franziskaner 1.20540643
#> grimbergen 1.78612198
#> guiness 0.31717542
#> hoegardeen 2.23601856
#> jupiler 2.41738081
#> kingfisher 1.17563178
#> latrappe 2.48017277
#> lindemanskriek 0.84132717
#> nicechouffe -0.20973451
#> pecheresse 2.73987210
#> sierranevada 2.12878315
#> tanglefoot 0.50802841
#> tauro 2.45585085
#> westmalle 1.86373375
#> amrut -1.63004033
#> ballantines -3.31173062
#> bushmills -1.09107572
#> chivas -1.97923449
#> dalmore -0.60822705
#> famousgrouse -1.43517709
#> glendronach -1.56712869
#> glenmorangie -1.46854222
#> highlandpark -2.62231929
#> jackdaniels -0.70483285
#> jb -2.24638367
#> johnniewalker -0.97832954
#> magallan -2.17603623
#> makersmark -1.23316404
#> oban -2.34336958
#> oldpotrero -0.37639098
#> redbreast -4.17639973
#> tamdhu -0.09818091
#> wildturkey -3.48193578
#> yoichi -2.32326072
#>
#> $f
#> type1 type2 type3 type4 type5 type6 type7 type8 type9 type10 type11
#> whisky whisky whisky whisky whisky whisky whisky whisky whisky whisky whisky
#> type12 type13 type14 type15 type16 type17 type18 type19 type20 type21 type22
#> whisky whisky whisky whisky whisky whisky whisky whisky whisky beer beer
#> type23 type24 type25 type26 type27 type28 type29 type30 type31 type32 type33
#> beer beer beer beer beer beer beer beer beer beer beer
#> type34 type35 type36 type37 type38 type39 type40
#> beer beer beer beer beer beer beer
#> Levels: beer whisky
#>
#> $colors_groups
#> [1] "#66C2A5FF" "#FC8D62FF"
#>
#> $colors_rows
#> [1] "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF"
#> [7] "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF"
#> [13] "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF"
#> [19] "#FC8D62FF" "#FC8D62FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF"
#> [25] "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF"
#> [31] "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF"
#> [37] "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF"
#>
#> $object
#> [1] "PCA"
#>
#> $axes
#> [1] 1
#>
#> $palette
#> function (n, transp = 0)
#> {
#> .pal_brewer(n, "Set2") %>% pal_alpha(transp = transp)
#> }
#> <bytecode: 0x5653330bb988>
#> <environment: namespace:Momocs>
#>
#> $method
#> [1] "LDAPCA"
#>
#> $mshape
#> NULL
#>
#> $cuts
#> NULL
#>
#> $eig
#> NULL
#>
#> $sdev
#> [1] 11.33732
#>
#> $rotation
#> LD1
#> PC1 -0.0021966326
#> PC2 -0.0004659024
#> PC3 0.0028778651
#> PC4 -0.0020601963
#> PC5 0.0016842402
#> PC6 -0.0008112386
#> PC7 -0.0006776597
#> PC8 -0.0004229307
#>
#> $LDs
#> LD1
#> PC1 -0.0021966326
#> PC2 -0.0004659024
#> PC3 0.0028778651
#> PC4 -0.0020601963
#> PC5 0.0016842402
#> PC6 -0.0008112386
#> PC7 -0.0006776597
#> PC8 -0.0004229307
#>
#> $baseline1
#> NULL
#>
#> $baseline2
#> NULL
#>
#> $links
#> NULL
#>
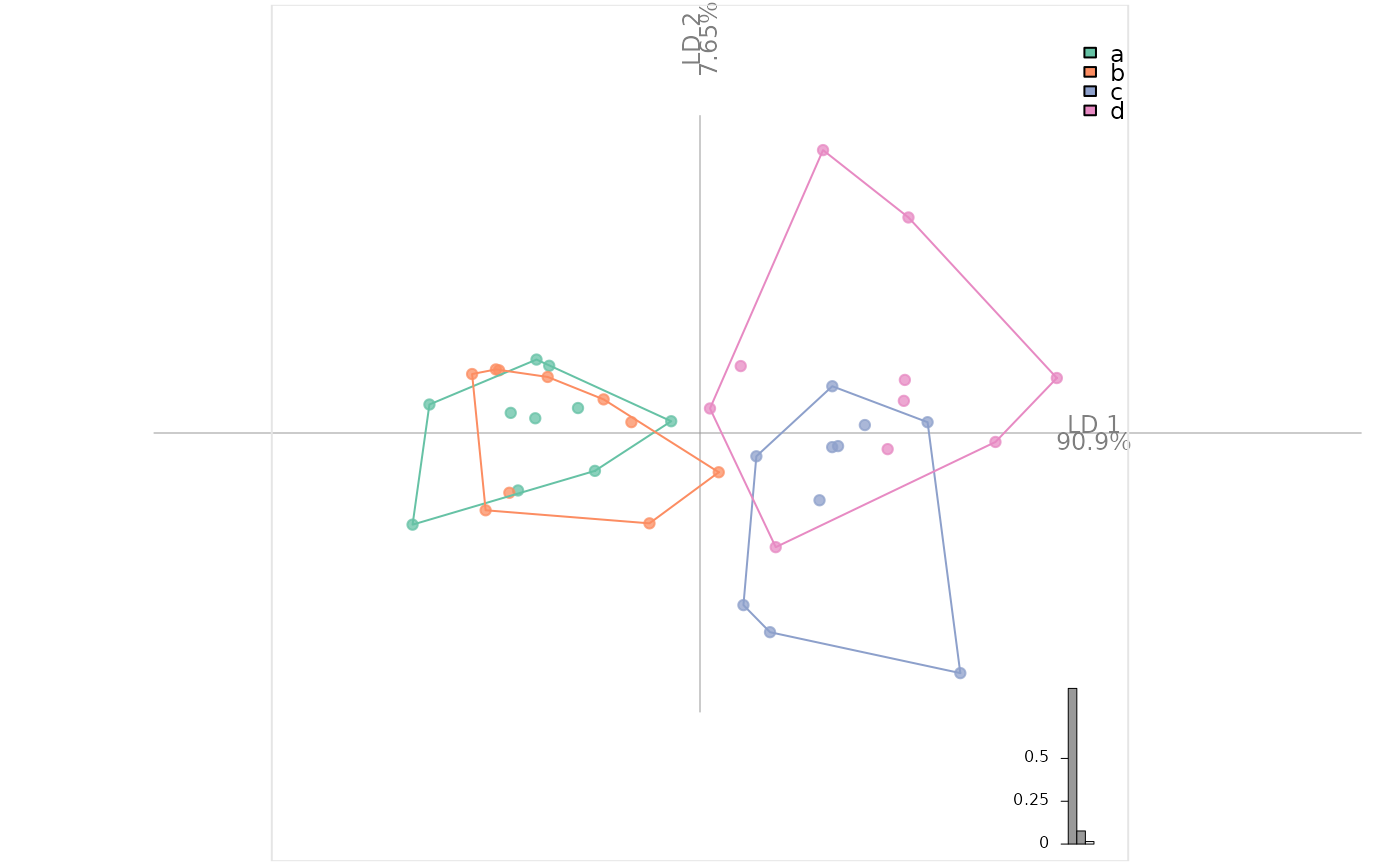
bot.f <- mutate(bot.f, plop=factor(rep(letters[1:4], each=10)))
bot.l <- LDA(PCA(bot.f), 'plop')
#> 8 PC retained
plot_LDA(bot.l) # will replace the former soon
#> $xy
#> LD1
#> brahma 2.06882655
#> caney 1.95733171
#> chimay 3.18567319
#> corona 1.91972111
#> deusventrue 1.51983847
#> duvel 3.25459981
#> franziskaner 1.20540643
#> grimbergen 1.78612198
#> guiness 0.31717542
#> hoegardeen 2.23601856
#> jupiler 2.41738081
#> kingfisher 1.17563178
#> latrappe 2.48017277
#> lindemanskriek 0.84132717
#> nicechouffe -0.20973451
#> pecheresse 2.73987210
#> sierranevada 2.12878315
#> tanglefoot 0.50802841
#> tauro 2.45585085
#> westmalle 1.86373375
#> amrut -1.63004033
#> ballantines -3.31173062
#> bushmills -1.09107572
#> chivas -1.97923449
#> dalmore -0.60822705
#> famousgrouse -1.43517709
#> glendronach -1.56712869
#> glenmorangie -1.46854222
#> highlandpark -2.62231929
#> jackdaniels -0.70483285
#> jb -2.24638367
#> johnniewalker -0.97832954
#> magallan -2.17603623
#> makersmark -1.23316404
#> oban -2.34336958
#> oldpotrero -0.37639098
#> redbreast -4.17639973
#> tamdhu -0.09818091
#> wildturkey -3.48193578
#> yoichi -2.32326072
#>
#> $f
#> type1 type2 type3 type4 type5 type6 type7 type8 type9 type10 type11
#> whisky whisky whisky whisky whisky whisky whisky whisky whisky whisky whisky
#> type12 type13 type14 type15 type16 type17 type18 type19 type20 type21 type22
#> whisky whisky whisky whisky whisky whisky whisky whisky whisky beer beer
#> type23 type24 type25 type26 type27 type28 type29 type30 type31 type32 type33
#> beer beer beer beer beer beer beer beer beer beer beer
#> type34 type35 type36 type37 type38 type39 type40
#> beer beer beer beer beer beer beer
#> Levels: beer whisky
#>
#> $colors_groups
#> [1] "#66C2A5FF" "#FC8D62FF"
#>
#> $colors_rows
#> [1] "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF"
#> [7] "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF"
#> [13] "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF" "#FC8D62FF"
#> [19] "#FC8D62FF" "#FC8D62FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF"
#> [25] "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF"
#> [31] "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF"
#> [37] "#66C2A5FF" "#66C2A5FF" "#66C2A5FF" "#66C2A5FF"
#>
#> $object
#> [1] "PCA"
#>
#> $axes
#> [1] 1
#>
#> $palette
#> function (n, transp = 0)
#> {
#> .pal_brewer(n, "Set2") %>% pal_alpha(transp = transp)
#> }
#> <bytecode: 0x5653330bb988>
#> <environment: namespace:Momocs>
#>
#> $method
#> [1] "LDAPCA"
#>
#> $mshape
#> NULL
#>
#> $cuts
#> NULL
#>
#> $eig
#> NULL
#>
#> $sdev
#> [1] 11.33732
#>
#> $rotation
#> LD1
#> PC1 -0.0021966326
#> PC2 -0.0004659024
#> PC3 0.0028778651
#> PC4 -0.0020601963
#> PC5 0.0016842402
#> PC6 -0.0008112386
#> PC7 -0.0006776597
#> PC8 -0.0004229307
#>
#> $LDs
#> LD1
#> PC1 -0.0021966326
#> PC2 -0.0004659024
#> PC3 0.0028778651
#> PC4 -0.0020601963
#> PC5 0.0016842402
#> PC6 -0.0008112386
#> PC7 -0.0006776597
#> PC8 -0.0004229307
#>
#> $baseline1
#> NULL
#>
#> $baseline2
#> NULL
#>
#> $links
#> NULL
#>
bot.f <- mutate(bot.f, plop=factor(rep(letters[1:4], each=10)))
bot.l <- LDA(PCA(bot.f), 'plop')
#> 8 PC retained
plot_LDA(bot.l) # will replace the former soon